Neural tube defects such as spina bifida, where a part of the spinal cord fails to close, may be caused by the same genes that lead to cleft lip and palate, according to a report last week (January 25) in Human Molecular Genetics. Scientists had previously thought the biological events surrounding neural tube and facial development were unrelated, but these new results, from both mouse models and human samples, link them in a way that may have to do with the timing and extent of activity among these genes.
What the researchers have done is “quite remarkable. . . . They suggest that these three genes form a network that’s a fundamental pathway for multiple morphogenetic [developmental] processes,” says Mary Marazita, the director of the University of Pittsburgh’s Center for Craniofacial and Dental Genetics. She was not involved in the study but has collaborated with several of its authors in the past. “I can’t wait to go back to some of our cleft data with this idea in mind.”
Cleft lip and palate are some of the most common birth defects, affecting about one in every 700 births. They occur when cells forming on either side of the head don’t grow all the way to the center of the face where they’d normally join. This can leave an opening in the infant’s upper lip or palate. The defects seem to run in families, and previous research has identified at least 50 parts of the genome related to an increased risk for clefting.
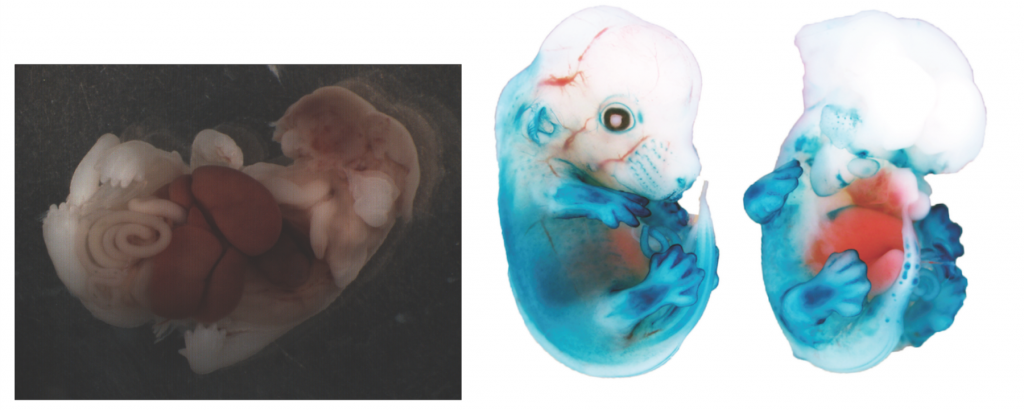
Youssef Kousa, now a clinical fellow at Children’s National Health System in Washington, DC, and the study’s lead author, was a graduate student breeding transgenic mice in hopes of understanding how one gene, interferon regulatory factor 6 (IRF6), influenced the formation of cleft lip and palate. But one of the embryos he bred had bizarre defects unrelated to the palate, so he brought it to his research mentor, Brian Schutte, a molecular geneticist at Michigan State University.
It was unlike anything Kousa had seen before. Among several striking deformities, the mouse embryo, which overexpressed the gene, was missing a part of its skull and brain, signifying a defect in the neural tube, a hollow cylinder that forms as the precursor for the brain and spinal cord. It wasn’t what the team planned to study, but it was “so amazing that we needed to pursue it,” Schutte tells The Scientist.
Nine years and close to 3,000 experiments later, the scientists identified a group of genes involved in the development of both the neural tube and the palate.
The team started by breeding more mice that overexpressed IRF6 and showed signs of neural tube defects. They hypothesized that if the hyperactive gene was causing the defect, crossing the first mice with ones that didn’t express IRF6 would even things out and create normal-looking mice. It did.
But they also found that both overexpression and under expression of IRF6 led to defects, albeit in different parts of the embryo. Too much IRF6 and the embryos displayed defects at the top of the neural tube, like the first embryo Kousa found. Too little and they had structural defects at the tail, which the team says may or may not be due to a defect in the neural tube.
The researchers did not look for clefting in these particular mice, although previous research has shown that 100 percent of mice that lack IRF6 develop these types of deformities.
“IRF6 homeostasis is what’s really important. You have to have that ‘magic Goldilocks amount.’ If you have too much it’ll cause one birth defect, if you have too little it’ll cause a different birth defect,” says Schutte.
The effects seem to be mediated by IRF6’s effect on two transcription factors—transcription factor AP2A (TFAP2A) and grainyhead like 3 (GRHL3)—which were already known to be play a role in the development of the lip and palate. Strangely, while they developed into different defects, both overexpression and underexpression of IRF6 led to a downregulation of GRHL3 and TFAP2A.
The researchers used “a huge number of animals . . . that contributes to the confidence of the result,” says Katherina Walz, director of the Division of Human Disease Modelling at the University of Miami, who was not involved in the study. She adds that the physical characteristics of the mice in the study were “really striking and impressive.”
To see if the IRF6 functioned similarly in humans, Schutte reached out to hospitals across the country that stored samples from patients with neural tube defects. These samples are typically hard to come by, as researchers estimate about 3.3 to 27.9 of every 10,000 births in the United States are affected, and many infants do not survive. In hundreds of samples, his team found two different mutations of IRF6 in two spina bifida patients, suggesting the gene is involved in human neural tube development as well that of the mouse.
None of the samples suggested that human patients simultaneously developed both neural tube and clefting defects. A previous epidemiological study even showed that patients with a cleft lip or palate were even less likely than the general population to have spina bifida. Yet, one patient in the study with spina bifida had the exact same mutation in IRF6 as a patient from an earlier study with a common form of cleft lip and palate, showing that the variant is tied to birth defects in different tissues that develop at different times and locations. “We absolutely do not understand that,” says Schutte.
The researchers believe the difference may lie in a patient’s genetic background or environmental exposures, but that’s a question for future work. The team hopes the work will lead to an improved ability to assess genetic risk factors for families, and of course, eventually, to prevention.
“As we think about gene discovery and identifying heritability, we might not need to look for more, more, more genes,” says Kousa. “The answer might be in some of the genes we already know pretty well.”