Some big ideas seem to appear out of nowhere, but in 2008 Chuan He deliberately went looking for one. The US National Institutes of Health had just launched grants to support high-risk, high-impact projects, and He, a chemist at the University of Chicago in Illinois, wanted to apply. But he needed a good pitch.
He had been studying a family of proteins that repair damaged DNA, and he began to suspect that these enzymes might also act on RNA. By a stroke of luck, he ran into molecular biologist Tao Pan, who had been investigating specific chemical marks, called methyl groups, that are present on RNAs. The pair worked in the same building at the University of Chicago, and began meeting regularly. From those conversations, their big idea took shape.
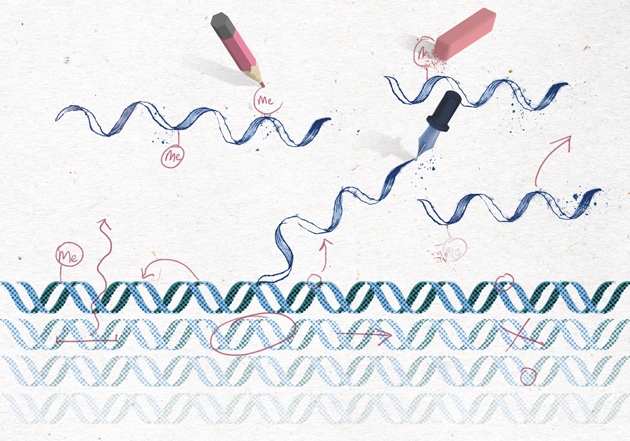
At the time, biologists were getting excited about the epigenome — the broad array of chemical marks that decorate DNA and its protein scaffold. These marks act like a chemical notation, telling the cell which genes to express and which to keep silent. As such, the epigenome helps to explain how cells with identical DNA can develop into the multitude of specialized types that make up different tissues. The marks help cells in the heart, for example, maintain their identity and not turn into neurons or fat cells. Misplaced epigenetic marks are often found in cancerous cells.
When He and Pan began working together, most epigenetic research focused on the tags associated with DNA and the histone proteins that it wraps around. But more than 100 different types of chemical mark had been identified on RNA, and nobody knew what they did. Some of the enzymes He was studying could strip off methyl groups, and He and Pan wondered whether one of them might work on RNA. If the marks could be reversed, they might constitute an entirely new way of controlling gene expression. In 2009, they got funding to hunt for reversible marks on RNA and the proteins that erase them.
Nine years later, such research has given birth to an ‘ome of its own, the epitranscriptome. He and others have shown that a methyl group attached to adenine, one of the four bases in RNA, has crucial roles in cell differentiation, and may contribute to cancer, obesity and more1, 2. In 2015, He’s lab and two other teams uncovered the same chemical mark on adenine bases in DNA (methyl marks had previously been found only on cytosine), suggesting that the epigenome may be even richer than previously imagined3. Research has taken off. “I think we’re approaching a golden age of epigenomics and epitranscriptomics,” says Christopher Mason, a geneticist at Weill Cornell Medical College in New York City. “We can actually start to see all these modifications that we knew have been there for decades.”
Marking the messenger
The governing rule of molecular biology — the central dogma — holds that information flows from DNA to messenger RNA to protein. Many scientists therefore viewed mRNA as little more than a courier, carrying the genetic information encoded in a cell’s nucleus to the protein factories in the cytoplasm. That’s one reason why few researchers paid much attention to the modifications made to mRNA.
They weren’t a secret, though. The mark that pushed He to the forefront of epitranscriptomics was first discovered on mRNA in 1974 (ref. 4). Fritz Rottman, an organic chemist at Michigan State University in East Lansing, was trying to understand the role of RNA in regulating gene expression when he stumbled across a methyl group on adenine. The modified base is called N6-methyladenosine, a mouthful that’s commonly shortened to m6A.
Rottman and his colleagues wrote that RNA methylation could be a way to select certain transcripts for translation into protein. “But that was all speculation,” says Karen Friderici, an author of the 1974 paper and a geneticist at Michigan State University. The team didn’t have a good way to investigate the mark’s true function. “It was the beginning of molecular biology. We didn’t have many of the tools that are available now,” she says.
More than three decades later, He and Pan found the tools still lacking. “It’s very difficult to actually study these modifications,” Pan says. It requires powerful mass spectrometry and high-throughput sequencing techniques.
Two members of He’s lab at the time, Ye Fu and Guifang Jia, pushed forward anyway, focusing on a protein called FTO, part of the family of methyl-stripping enzymes that He’s group had been studying. Fu and Jia thought that it might remove methyl groups from RNA, but they struggled to identify its target. Fu and his colleagues began to synthesize snippets of RNA that contained different modifications, to determine whether FTO could remove them. It was slow going. Over the course of three years, the team faced a string of failures, “I almost thought I would never find the function,” Fu says.
Finally, in 2010 the team decided to test FTO’s activity on m6A — the methylated adenine. The mark disappeared. The team had shown for the first time that RNA methylation was reversible5, just like the marks found on DNA and histones. To He, it seemed like proof of an RNA-based system of gene regulation.
Evidence mounts
He’s group wasn’t the only one thinking about m6A. In 2012, two teams of researchers independently published the first maps of where m6A appears6, 7. The studies revealed more than 12,000 methylated sites on mRNAs originating from about 7,000 genes. “After years in the dark, we were instantly facing a wide vista,” wrote Dan Dominissini, an author of one of the studies, in an essay in Science8.
The maps showed that the distribution of m6A is not random. Its location suggested that the mark might have a role in alternative splicing of RNA transcripts, a mechanism that allows cells to produce multiple versions of a protein from a single gene.
Over the past few years, researchers have identified some of the machinery involved in regulating these marks. Each requires a writer to place it, an eraser to remove it and a reader to interpret it (see ‘Reading, writing and regulation’). As the identities of these proteins emerged, scientists have come to understand that m6A affects not only RNA splicing, but also translation and RNA stability…..