The completion of the human genome sequence more than a decade ago was an indisputable triumph for biomedical research. And more recently, efforts such as the Encyclopedia of DNA Elements (ENCODE) project have sought to expand knowledge of functional elements within the genome.
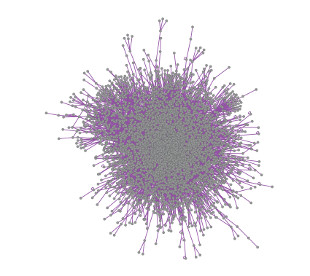
But truly connecting genotype to phenotype will require a comprehensive view of how the protein products of genes operate and interact. Researchers at the Dana-Farber Cancer Institute’s Center for Cancer Systems Biology and their colleagues have produced a new human interactome map, reported today (November 20) in Cell. The map is based on a systematic screen of 13,000 human proteins that uncovered 14,000 pairwise interactions.
This nine-year project likely represents about 5 percent to 10 percent of all the protein-protein interactions that exist, according to study coauthorFritz Roth of the University of Toronto. While still limited in scope, it is at least a five-fold improvement over previous interactome maps, Roth added.
“This is a long road, and we’ve never had a human interactome project to go with the Human Genome Project,” he said. “But I think people are starting to appreciate that the genome is the beginning of the story it’s a parts list in an alien language that we’re starting to figure out.”
To identify these interactions, the researchers used a high-throughput yeast two-hybrid approach, in which 82 million protein pairs were each tested four times in two different configurations for their ability to activate a reporter gene in yeast. The researchers also validated selected interactions using three independent methods, testing whether the protein pairs could reconstitute the parts of a fluorescent protein or a membrane-bound protein complex in mammalian cells, plus used an in vitro method.
The team compared its results to a list of protein interactions supported by multiple pieces of evidence garnered from a literature search in 2013. The researchers found that their systematic strategy picked up a large swath of interactions that were missed by individual studies.
“This kind of centralized approach has a much higher likelihood of finding interactions throughout the human proteome, rather than just finding interactions of the specific proteins that people have studied because of a disease process or because of the specific cellular function that they’re interested in,” said Stanley Fields of the University of Washington. Fields, who pioneered the use of the yeast two-hybrid system, was not involved in the present study, but served on the advisory board for a National Institutes of Health (NIH) grant that partially funded the research.
Notably, the new interactome map lends support to a long-held suspicion that proteins implicated in cancer participate in disproportionate numbers of interactions with other proteins. As scientists sequence tumor genomes, more extensive knowledge of these protein-protein interactions could help to distinguish “driver” mutations that cause cancer from “passenger” mutations that are simply along for the ride.
“Our goal is to help facilitate the expansion and robustness of the human interactome to the point that it can really provide insight into every chronic disease,” said Joseph Loscalzo of Harvard Medical School, a cardiovascular researcher and long-time proponent of “network medicine” who has collaborated with study coauthor Albert-László Barabási, but was not involved in the present work.
While this latest map is a valuable resource, it provides a static view of the proteome, said Loscalzo: “Looking at dynamic changes will be another important part of this it would also be useful to look at adaptive responses of the proteome to stresses in the environment.”
Roth and his collaborators are already at work on the next interactome map, which will expand the screen to 17,000 proteins. While large, this expanded map will still be far from comprehensive. “One thing we know is that not every interaction can be detected by every assay, so it’s unlike genome sequencing,” he said. “It’s an asymptotic problem.”
Fields suggested that future efforts might consider tissue-specific proteins and posttranslational modifications that affect protein-protein interactions, in addition to proteins produced from alternatively spliced transcripts. “Ultimately, the interaction maps are going to be way more complicated than just the genomic sequence has proven to be,” he said, “but you have to start somewhere.”