A newly developed method called DNA microscopy can accurately pinpoint the location of mRNA molecules within a cell and produce a visual map of the transcripts—all without actually using a microscope.
The technique, published in Cell today (June 20), works by first converting selected RNAs in a cell culture to cDNAs, then labeling each molecule with a unique stretch of nucleotides. Those labels then amplify themselves, and when they expand to the point of contacting another cDNA’s labels, the connection itself gets a unique nucleotide tag. The closer any two cDNAs are, the more connection tags they’ll generate.
As Science describes it, “To count the labels, the researchers grind up the cells and analyze the DNA they contain. A computer algorithm can then infer the original positions of the DNA molecules to generate an image.”
“The first time I saw a DNA microscopy image, it blew me away,” coauthor Aviv Regev, a computational and systems biologist at the Broad Institute, tells The New York Times.
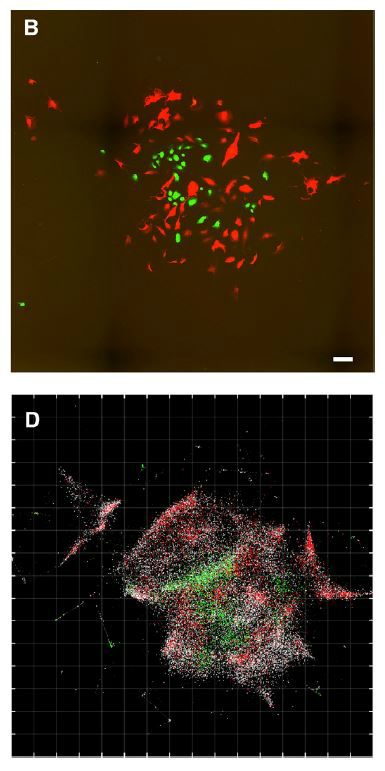
The researchers compared their computer-generated images with true microscopy images of the same cells, and found that while DNA microscopy wasn’t as crisp, the locations of the selected mRNAs were accurately plotted. “[O]ne key weakness of DNA microscopy remains the resolution of empty space,” which could distort the map when RNA molecules are far apart, Regev and her colleagues write in their study, “and future work will be needed to eliminate this obstacle to produce high-quality reconstructions of samples over large lengths where there are gaps in molecular density.”