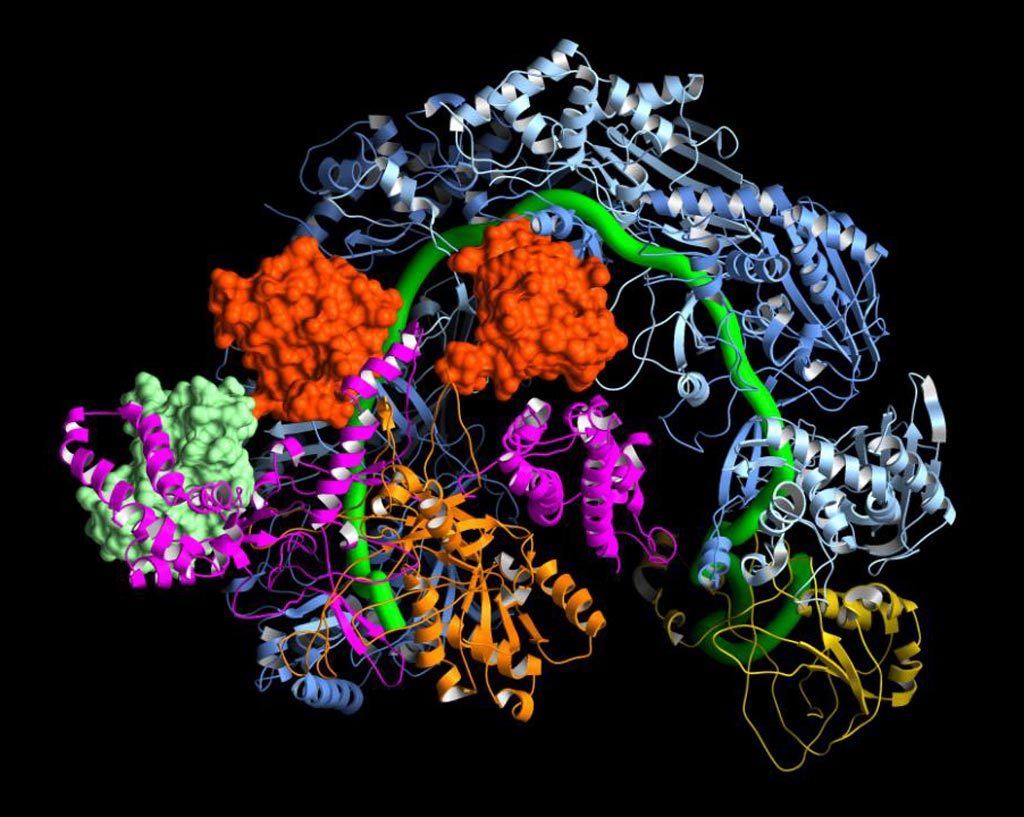
Cryo-electron microscopy (Cryro-EM) molecular mapping was used to determine how the bacterial gene editing complex CRISPR/Cas9 interacts with viral anti-CRISPR proteins.
CRISPRs (clustered regularly interspaced short palindromic repeats) are segments of prokaryotic DNA containing short repetitions of base sequences. Each repetition is followed by short segments of “spacer DNA” from previous exposures to a bacterial virus or plasmid. CRISPRs are found in approximately 40% of sequenced bacteria genomes and 90% of sequenced archaea. CRISPRs are often associated with cas genes that code for proteins related to CRISPRs.
Since 2013, the CRISPR/Cas system has been used in research for gene editing (adding, disrupting, or changing the sequence of specific genes) and gene regulation. By delivering the Cas9 enzyme and appropriate guide RNAs (sgRNAs) into a cell, the organism’s genome can be cut at any desired location. The conventional CRISPR/Cas9 system is composed of two parts: the Cas9 enzyme, which cleaves the DNA molecule and specific RNA guides that shepherd the Cas9 protein to the target gene on a DNA strand. Prokaryotes evolved CRISPR-mediated adaptive immune systems for protection from viral infection, and viruses have evolved diverse anti-CRISPR (Acr) proteins that subvert these immune systems.
Investigators at the Scripps Research Institute reported in the March 23, 2017, issue of the journal Cell that they had used Cryo-EM to determine the structure of the Pseudomonas aeruginosa CRISPR complex bound to two different Acr proteins, AcrF1 and AcrF2, at an average resolution of 3.4 Angstroms.
Researchers have historically relied on NMR and X-ray diffraction techniques to determine the structures of molecular complexes and proteins that play a role in the causes of various disease states. Structural information about a variety of medically important proteins and drugs has been obtained by these methods. Cryo-EM is a complementary analytical technique that provides near-atomic resolution without requirements for crystallization or limits on molecular size and complexity imposed by the other techniques. Cryo-EM allows the observation of specimens that have not been stained or fixed in any way, showing them in their native environment while integrating multiple images to form a three-dimensional model of the sample.
The molecular structure obtained during this study explained the mechanism for immune system suppression, and structure-guided mutations showed that the Acr proteins bound to residues essential for crRNA-mediated detection of DNA.
“These findings are important because we knew that anti-CRISPR proteins were blocking bacterial defenses, but we had no idea how,” said senior author Dr. Gabriel C. Lander, assistant professor of integrative structural and computational biology at the Scripps Research Institute. “This system can quickly read through massive lengths of DNA and accurately hit its target. If the CRISPR complex identifies a viral DNA target, the surveillance machine recruits other molecules to destroy the virus’s genome. These anti-CRISPR proteins keep the bacteria from recognizing the viral DNA. CRISPR systems cannot escape from these anti-CRISPR proteins without completely changing the mechanism they use to recognize DNA.”
“Although CRISPR/Cas9 is the “celebrity” CRISPR system, there are 19 different types of CRISPR systems, each of which may have unique advantages for genetic engineering. They are a massive, untapped resource,” said Dr. Lander. “The more we learn about the structures of these systems, the more we can take advantage of them as genome-editing tools.”