Abstract
Satellites are mobile genetic elements that are dependent upon the replication machinery of their helper viruses. Bacteriophages have provided many examples of satellite nucleic acids that utilize their helper morphogenic genes for propagation. Here we describe two novel satellite-helper phage systems, Mulch and Flayer, that infect Streptomyces species. The satellites in these systems encode for encapsidation machinery but have an absence of key replication genes, thus providing the first example of bacteriophage satellite viruses. We also show that codon usage of the satellites matches the tRNA gene content of the helpers. The satellite in one of these systems, Flayer, does not appear to integrate into the host genome, which represents the first example of a virulent satellite phage. The Flayer satellite has a unique tail adaptation that allows it to attach to its helper for simultaneous co-infection. These findings demonstrate an ever-increasing array of satellite strategies for genetic dependence on their helpers in the evolutionary arms race between satellite and helper phages.
Introduction
The virosphere contains many instances of virus-like mobile genetic elements (MGEs). Satellites are virus-like MGEs that depend on another virus for replication, and they are classified on the basis of this dependence [1]. Satellite viruses are dependent on helper viruses for genome replication, whereas satellite nucleic acids depend on helper viruses for transmission via encapsidation using helper-derived capsids [2]. No satellite viruses infecting bacterial hosts have been reported to date, but several families of satellite nucleic acids have been described and are commonly referred to as phage satellites [3].
Bacteriophage systems provided some of the first examples of satellite nucleic acids [4]. Enterobacteria phage P4 infects Escherichia coli cells and can integrate in the bacterial chromosome or exist as a plasmid, but requires the morphogenic genes of a co-infecting Enterobacteria phage P2 in order to encapsulate and lyse the cell [5]. Many other satellite nucleic acids have been described infecting a diverse range of bacterial hosts. Like P4-like satellites, phage-inducible chromosomal islands (PICIs) and PICI-like elements (PLEs) are widely disseminated satellites that reside in the bacterial chromosome and depend on helper viruses for encapsidation and lysis [6,7,8]. These satellite nucleic acids often encode genes that confer adaptive advantage to their bacterial host and typically interfere with helper phage replication [6]. Satellites associated with lytic phages, such as the Vibrio cholerae PLE, completely restrict production of their helper phage, effectively functioning as abortive infection systems [9].
All known phage satellites integrate into the host chromosome and replicate using satellite-encoded replicases [8]. Phage satellites use several strategies to divert encapsidation from helper phages. Enterobacteria phage P4, for instance, encodes a scaffolding protein (Sid) that redirects P2 capsid assembly, generating smaller capsids that cannot physically accommodate the P2 genome. P4 DNA is then packaged into the smaller capsids by the P2 terminase, which recognizes cos sites in the P4 chromosome [10]. Prototypical PICIs, like the staphylococcal pathogenicity islands (SaPIs), also use scaffolding proteins to generate smaller capsids, but display different packaging strategies. Some SaPIs contain cos sites and make direct use of the helper’s terminase for packaging, relying primarily on size exclusion to prioritize their preferential packaging [11]. Other SaPIs encode a small terminase subunit (TerS) to redirect packaging to the SaPI-specific pac sites and an interference protein to inactivate the phage TerS [3]. Proteobacterial PICIs encode a protein (Rpp) that binds the phage TerS, promoting its specific recognition of PICI cos sites [12].
Recently, a new class of PICI has been reported [13]. In contrast with previously described PICIs, capsid-forming PICIs (cf-PICIs) encode all the functional components for capsid formation (major capsid protein, head maturation protease, portal protein and head-tail connectors) as well as packaging (large and small terminase subunits and HNH endonuclease). These PICIs, which are widely distributed across Bacteria, therefore depend only on their helper phage for the production of tails [13].
Here, we report the isolation and characterization of a novel group of satellite phages infecting Streptomyces species that encode a full repertoire of capsid formation and packaging genes. These satellites present no significant homology with canonical or capsid-forming PICIs, PLEs or P4-elements, and possess several distinctive features: they parasitize virulent Streptomyces phages, their genomes contain direct terminal repeats (DTRs) and they present anomalously low %GC content matching their helper’s. Furthermore, our results indicate that loss of the chromosomal integration module in one of these satellites has been compensated by satellite acquisition of specialized tail components that appear to attach to the helper phage to facilitate co-infection via simultaneous entry into the host. To our knowledge, this is the first description of a virulent satellite associated with a virulent helper.
Methods
Isolation, characterization, and DNA sequencing
The MulchMansion/MulchRoom and the MindFlayer/MiniFlayer systems were isolated, respectively, from soil samples collected in St. Louis, MO (USA) and Poolesville, MD (USA), using previously reported phage isolation methods [14]. Soil samples were first suspended in phage buffer (10 mM Tris pH7.5, 10 mM MgSO4, 1 mM CaCl2, 68.5 mM NaCl). The resulting suspensions were spun to pellet soil and the supernatant filtered using a ø0.22 μm filter. The filtrate was then added to tryptic soy (TS) soft agar (BD, Sparks, MD, USA) with Streptomyces lividans JI 1326 (MulchMansion/MulchRoom) or Streptomyces mirabilis NRRL B-2400 (MindFlayer/MiniFlayer) and overlayed onto nutrient agar plates (BD Difco, Sparks, MD) supplemented with 10 mM MgCl2, 8 mM Ca(NO3)2 and 0.5% glucose (NA+). Plates were incubated at 30 °C for 1 to 2 days. A minimum of three rounds of plaque purification was carried out as previously described [14]. In short, isolated plaques were picked into phage buffer and serially diluted, then 10 µL of each dilution was combined with a 48 h culture of host, incubated 10 min at room temperature, combined with TS soft agar and plated on NA+ plates.
Crude stock lysate was harvested as described before [14] from plates demonstrating near-confluent lysis of S. mirabilis post infection. Plates were flooded with 5–8 mL phage buffer. The buffer (MulchMansion/MulchRoom) or buffer and soft agar overlay (MindFlayer/MiniFlayer) was collected and centrifuged 20 min at 2,500x g, and the supernatant passed through a ø0.22 µm filter. To screen for potential lysogens, MindFlayer crude lysate was serially diluted and the dilutions spotted on a freshly prepared plate of S. mirabilis and incubated for seven days at 30 °C.
DNA for the MindFlayer/MiniFlayer and MulchMansion/MulchRoom systems was isolated using the Promega Wizard DNA purification system on freshly prepared high-titer lysates, as reported previously [15]. Sequencing of MulchMansion/MulchRoom was performed by the McDonnell Genome Institute at Washington University with a NovaSeq 6000 (Illumina, USA) using High-Throughput Library Preparation Kit Standard PCR Amp Module (KAPA Biosystems, Boston, USA) and 150 × 2 reads. Sequencing of MindFlayer/MiniFlayer was completed by the Pittsburgh Bacteriophage Institute with the MiSeq (Illumina; v3 reagents) sequencing platform using the NEB Ultra II Library Kit and 150-base single-end reads. For all assemblies, raw sequencing reads were assembled using Newbler v2.9 or CLC Genomics Workbench NGS de novo assembler v6 with default settings. Genome completeness and termini were determined using Consed v. 29 [16, 17].
Genome assembly and annotation
Genome annotation was performed using DNA Master v5.23.6 [18], using the embedded Glimmer v.3.02b [19] and GeneMark v.4.28 [20] for protein-coding gene calling and Aragorn v1.2.41 for tRNA gene prediction [21]. Protein-coding gene calls were manually refined based on their proximity and directionality, the presence of putative ribosome binding sites and sequence similarity to previously annotated genes. Protein coding genes were functionally annotated through homology search with BLASTp [22] and HHPred v57c87 [23].
Electron microscopy
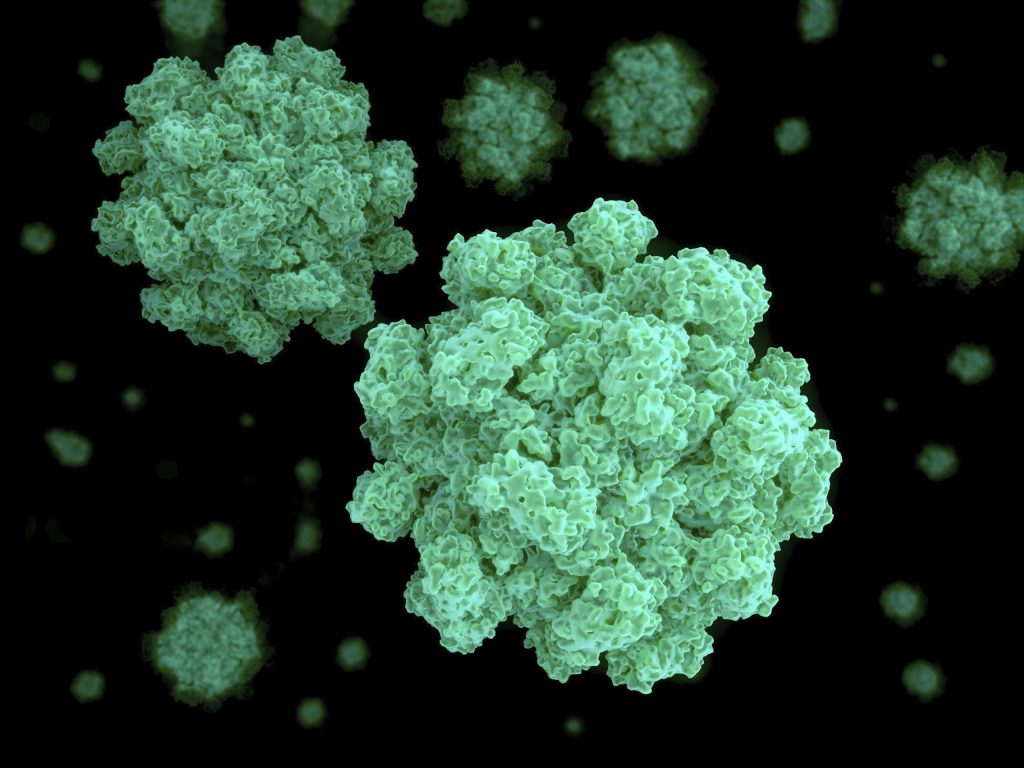
The MindFlayer/MiniFlayer system was imaged on a Morgagni M268 Transmission Electron Microscope (FEI, Hillsboro, IL, USA) equipped with an Orius CCD camera (Gatan Inc., Pleasanton, CA, USA). 10 µL of crude lysate was placed on to 200 mesh formvar-covered, carbon-coated copper grids (EMS, Hatfield, PA, USA), incubated for 1 min, briefly rinsed with ultra-pure water, then stained with 2% uranyl acetate for 2 min. Grids were scanned to record the number and position of associated satellites for randomly selected helper virions, as well as images taken for analysis. Morphological measurements were taken in Fiji [24] and all values are expressed as mean ± standard deviation. For certain analyses noted in the results, we also used “picked” plaque samples that are prepared by placing 50 µL of phage buffer directly onto a plaque and incubating for 30 s before pipetting up the liquid, followed by staining methods described above for the crude lysate.
Comparative genomics and phylogenetic analysis
Gene content similarity among phage genomes was computed using the PhagesDB service [25]. Genome map figures were generated with Easyfig [26], using an e-value threshold of 10–10 for tBLASTx and considering only hits above 25% identity. To perform phylogenetic inference, a set of phage genomes potentially related to bacteriophages MiniFlayer or MulchRoom was compiled (Table S1). The set was obtained through a combination of BLASTp and tBLASTn searches. tBLASTn was used to identify related phage genomes, by querying the NCBI GenBank database with all the proteins encoded by each satellite. The search was restricted to viruses (taxonomy ID 10239). Any genome containing at least one hit with E-value lower than 10–10 and query coverage larger than 75% was included (Tables S2, S3). To identify putatively related satellites integrated in bacterial chromosomes, BLASTp was performed against the proteomes of satellites reported in Ref. [8] using the same limiting thresholds as in the tBLASTn search (Tables S4, S5). The nucleotide sequences of satellites containing at least one hit were extracted from the GenBank genome records of the hosts (Table S6) and added to the set of potentially related phages. For reference, a small set of representatives of classic helper-satellite systems was also included (Table S7).
ViPhy [27] was used to obtain a phylogenetic distance matrix of phage genomes. We then performed agglomerative clustering using scikit-learn to produce a diverse set of 100 phages. A phylogenetic tree for these 100 phages was generated using the VICTOR web service [28]. Intergenomic distances were based on protein sequence distances, computed with the Genome-BLAST Distance Phylogeny (GBDP) method, using the settings recommended for prokaryotic viruses [28] and applying the d6 distance formula [29]. A balanced minimum evolution tree was produced with FASTME from the intergenomic distances [30], using 100 pseudo-bootstrap replicates to obtain branch support values. The phylogenetic tree was annotated and visualized with the iTOL web service [31]. Phage lifestyle (temperate/virulent) was predicted using the BACterioPHage LIfestyle Predictor (BACPHLIP) classifier [32]. The %GC content of phage genomes was computed using BioPython [33]. Lifestyle and %GC content were integrated for visualization in iTOL.
Genome sequence processing and codon usage bias analysis
Codon usage adaptation to tRNA gene pools was estimated by computing the tRNA Adaptation Index (tAI), as described in Ref. [34] (https://github.com/ErillLab/SPA). The genome sequences of the BE cluster phages and their hosts were downloaded from NCBI (Table S10). Both tRNAscan and Aragorn were used to predict tRNAs in downloaded genomes [21, 35]. Results were filtered for duplicates. When the same tRNA was predicted by both tRNAscan and Aragorn (same anticodon, maximum difference in location of 10 bp), the Aragorn tRNA location was kept. For tRNA genes with a predicted CAT anticodon, the tRNAscan tRNA call was kept. Whenever a tRNA was undetermined by tRNAscan, the Aragorn tRNA call was kept [36].
All the code used in this work can be found in a dedicated GitHub repository (https://github.com/ErillLab/SPA).
Results
Identification of a Streptomyces satellite-helper system through DNA sequencing
Streptomyces phages MindFlayer/MiniFlayer and MulchMansion/MulchRoom were isolated from soil samples collected in Poolesville, MD and St. Louis, MO (USA) using, respectively, S. mirabilis and S. lividans as isolation hosts. After 48 h at 30 °C on their isolation host, MindFlayer/MiniFlayer formed clear plaques with diameters of 1 to 5 mm (Supplementary Fig. 1A). Small plaques were homogeneously round, whereas larger plaques presented irregular shapes. MulchMansion/MulchRoom formed small (ø0.1 mm), sometimes cloudy plaques (Supplementary Fig. 1B). Following DNA purification, sequencing was performed using the Illumina MiSeq sequencing platform. Read assembly of MindFlayer/MiniFlayer resulted in two well-defined contigs corresponding to Streptomyces phages MindFlayer (130,258 bp) and MiniFlayer (17,449 bp), with 73% of the 297,433 reads mapping to the smaller contig. Read assembly of MulchMansion/MulchRoom also yielded two well-defined contigs corresponding to Streptomyces phages MulchMansion (134,105 bp) and MulchRoom (14,787 bp), with 78% of the reads mapping to the smaller contig. In all cases, the assembled chromosomes were found to be linear with direct-terminal repeats (DTRs) (Table 1). Plaque purification was attempted for the MindFlayer/MiniFlayer system, but was only successful for the larger phage (MindFlayer) as confirmed by PCR. Based on the sequencing read asymmetry, consistent with known satellite-helper systems [7], and the inability to purify the smaller phages, these two groups of phages were tentatively considered satellite-helper systems, hereafter referred to as the Mulch (MulchMansion/MulchRoom) and Flayer (MindFlayer/MiniFlayer) systems…