Quantitation of metabolic pathway regulation
Although metabolic biochemical pathways are well understood, less is known about precisely how reaction rates or fluxes through the various enzymes are controlled. Hackettet al. developed a method to quantitate such regulatory influence in yeast. They monitored concentrations of metabolites, enzymes, and potential regulators by LC-MS/MS (liquid chromatography–tandem mass spectrometry) and isotope ratio measurements for 56 reactions, over 100 metabolites, and 370 metabolic enzymes in yeast in 25 different steady-state conditions. Bayesian analysis was used to examine the probability of regulatory interactions. Regulation of flux through the pathways was predominantly controlled by changes in the concentration of small-molecule metabolites rather than changes in enzyme abundance. The analysis also revealed previously unrecognized regulation between pathways.
INTRODUCTION
Metabolism is among the most strongly conserved processes across all domains of life and is crucial for both bioengineering and disease research, yet we still have an unclear understanding of how metabolic rates (fluxes) are determined. Qualitatively, this deficiency involves missing knowledge of enzyme regulators. Quantitatively, it involves limited understanding of the relative contributions of enzyme and metabolite concentrations in controlling flux across physiological conditions. Addressing these gaps has been challenging because in vitro biochemical approaches lack the physiological context, whereas models of cellular metabolic dynamics have limited capacity for identifying or quantitating specific regulatory events because of overall model complexity.
RATIONALE
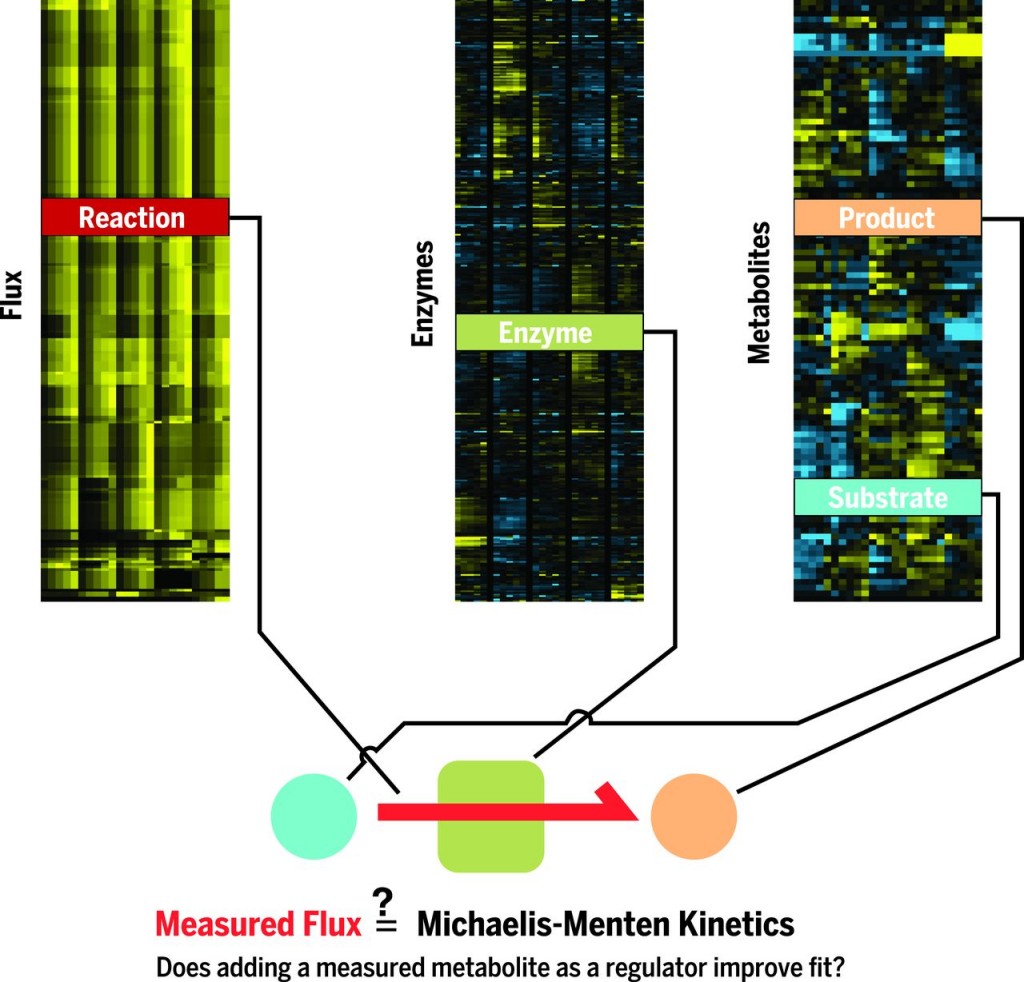
Flux through individual metabolic reactions is directly determined by the concentrations of enzyme, substrates, products, and any allosteric regulators, as captured quantitatively by a Michaelis-Menten–style reaction equation. Analogous to how experimental variation of reaction species in vitro allows for the inference of regulators and reaction equation kinetic parameters, physiological changes in flux entail a change in reaction species that can be used to determine reaction equations on the basis of cellular data. This requires measurement across multiple biological conditions of flux, enzyme concentrations, and metabolite concentrations. We reasoned that chemostat cultures could be used to induce predictable and strong flux changes, with changes in enzymes and metabolites measurable by proteomics and metabolomics. By directly relating cellular flux to the reaction species that determine it, we can carry out regulatory inference at the level of single metabolic reactions by using cellular data. An important benefit is that the physiological significance of any identified regulator is implicit from its role in determining cellular flux.
RESULTS
Here we introduce systematic identification of meaningful metabolic enzyme regulation (SIMMER). We measured fluxes, and metabolite and enzyme concentrations, in each of 25 yeast chemostats. For each of 56 reactions for which the flux, enzyme, and substrates were measured, we determined whether variation in measured flux could be explained by simple Michaelis-Menten kinetics. We also evaluated alternative models of each reaction’s kinetics that included a suite of allosteric regulators drawn from across all organisms. For 46 reactions, we were able to identify a useful kinetic model, with 17 reactions not including any regulation and 29 reactions being regulated by one to two allosteric regulators. Three previously unrecognized cross-pathway regulatory interactions were validated biochemically. These included inhibition of pyruvate kinase by citrate and inhibition of pyruvate decarboxylase by phenypyruvate. These metabolites accumulated and thereby curtailed glycolytic outflow and ethanol production in nitrogen-limited yeast. For well-supported reaction forms, we were able to determine the extent to which nutrient-based changes in flux were determined by changes in the concentrations of individual reaction species. We find that substrates are the most important determinant of fluxes in general, with enzymes and allosteric regulators having a comparably important role in the case of physiologically irreversible reactions.
CONCLUSION
By connecting changes in flux to their root cause, SIMMER parallels classic in vitro approaches, but it allows simultaneous testing of numerous regulators of many reactions under physiological conditions. Its application to yeast showed that changes in flux across nutrient conditions are predominantly due to metabolite, not enzyme, levels. Thus, yeast metabolism is substantially self-regulating.