Imaging
Life-science imaging broke barriers this year, as scientists built upon microscopy approaches to peer ever deeper into living tissues.
In October, Purdue University’s Ji-Xin Cheng and colleagues reported they had greatly increased the speed of collecting images from minutes to seconds using in vivo vibrational spectroscopic imaging, a technique that obviates the need for fluorescence. The key improvement was eliminating the need for a spectrometer, which collects the vibrational signature of molecules within a sample that are excited by light. Instead, Cheng’s team color-coded the photons before they went into the tissue.
“The idea is that before we send the photons into the tissue, we code every color with a distinct megahertz frequency,” Cheng told The Scientist at the time. “In this way, we can collect the diffuse photons within tens of microseconds and retrieve the spectrum through a one-to-one correspondence between the coding frequency and the color of light.”
“This is good progress toward making this technique more practical,” said bioengineering professor Stephen Boppart, who develops novel imaging modalities at the University of Illinois and was not involved in the work. “The authors have made the acquisition faster, allowing image collection in vivo and in highly photon scattering tissues.”
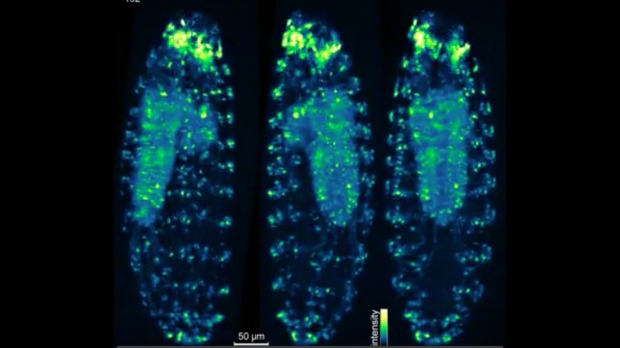
That same month, a team led by Philipp Keller of the Janelia Research Campus in Ashburn, Virginia, demonstrated its scope could visualize an entire, non-transparent animal in three dimensions and on biologically relevant time scales. The researchers imaged the development of the fruit fly nervous system, which ends up with a total of 10,000 cells. “This is far larger than the 302 neurons of C. elegans, the only other animal whose entire nervous system has been imaged at the single-cell level,” The Scientist noted in October.
The Keller team’s scope includes four objective lenses, whose images are combined, yielding 3.2 gigabytes of data per second. “Once we’ve registered and deconvolved the images, we get a very crisp version of the data,” Keller said in a press release. “Right now it’s our most advanced microscope for imaging, and we are certainly going to take advantage of it.”
Optogenetics
In November, Edward Boyden of MIT and Karl Deisseroth of Stanford University were honored for their work in developing optogenetics, which allows for the manipulation of neurons via light. Scientists have continued to improve upon the technique, notably, by combining the optogenetic control of neurons with the optogenetic readout of their activities.
One month before, at the Society for Neuroscience meeting in Chicago, Deisseroth and others presented updates on all-optical electrophysiology manipulating and recording neurons with light. Adam Cohen of Harvard and his team developed a reporter that when introduced into cells glows in the infrared upon changes in voltage. Along with Boyden, Cohen put the voltage indicator into cells along with a membrane channel that responds to blue light allowing the researchers to turn on cells with blue light and record their activities with infrared.
“The dream is to start doing recordings in intact neural circuits to see how a single neuron takes its inputs and combines those inputs and then decides whether or not to fire,” Cohen told The Scientist in October.
Also this year, researchers discovered a channelrhodopsin in algae that suppresses neuronal activity far faster than the engineered channels previously developed. This opens up options for far more precise inhibition of cell firing in optogenetics.
Scientists also worked out a way to give immediate feedback to neurons under optogenetic control, to maintain their activity at a desired state. This “neuro thermostat” can hold cell’s firing rate constant for 24 hours. “Just as a thermostat responds to temperature, if the firing rate deviates from a desired rate programmed into a computer receiving the electrode signals, the light intensity is immediately adapted to compensate for the change,” The Scientist reported in October.
Single-cell analyses
By parsing the unique aspects of individual cells, scientists this year were able to identify what appears to be a new bacterial phylum and detect the rarest of cell types within the mouse intestine. Single-cell analytics has boomed in recent years as protocols have become higher in throughput and precision.
Sorting or analyzing individual cells has become ever more practical as scientists have adapted microfluidics in clever ways, from being able to image cell deformities one-by-one to grabbing a snapshot of a cell’s molecular composition. Single-cell transcriptomics made a considerable leap forward this year by vastly expanding the number of cells that can be scrutinized in parallel, from about 100 to many thousands and up. Marc Kirschner and Steve McCarroll, both of Harvard, independently developed microfluidics devices that capture a single cell in a droplet along with all the reagents to prep the cell’s cDNA library.
“The innovation was using an aqueous-oil emulsion,” for the test-tube-in-a-droplet, Donald Zack of Johns Hopkins Hospital told The Scientist over the summer. “Instead of needing a well or a little spot for each cell, now they could, by many orders of magnitude, increase throughput.”
Single-cell analytics is not chained to microfluidic devices. For instance, researchers used a teeny, tiny capillary needle, similar to those used in in vitro fertilization, to suck out the contents of an individual plant cell. The material was then shot into a mass spectrometer to shed some light on the cell’s composition. Although much of the spectrometry readout are unidentified peaks, the new protocol “is very useful to the research community to move this type of analysis forward,” Sixue Chen of the University of Florida toldThe Scientist this fall.
CRISPR
The gene-editing tool known familiarly as CRISPR is the gift that keeps on giving. Scientists are continually demonstrating novel applications, from controversial tweaks to human embryos to restoring (in vitro) a protein deficient in a human disease to designing antimalaria mosquitoes.
While many researchers are putting CRISPR to use, others are focused on improving the technique itself. In a marriage of CRISPR and optogenetics, scientists built a system that assembles two pieces of the Cas9 enzyme—the CRISPR element responsible for snipping target DNA—upon exposure to blue light. When the light is off, the pieces break apart and CRISPR activity stops. “Such an on/off-switching property of [photoactivatable Cas9 nuclease] is the most important breakthrough previously unattainable,” University of Tokyo chemist Moritoshi Sato, who helped developed the technology, told The Scientist in June.
Cas9 is most frequently the object of CRISPR refinements. Earlier this month, for instance, MIT’s Feng Zhang and colleagues presented a refined Cas9 enzyme. They swapped three positively charged amino acids with charge-neutral alanine. The change “hindered Cas9’s ability to bind off-target DNA sites without significantly affecting the correct edits,” The Scientist reported.
Earlier in the year, Zhang’s team made another adjustment to the Cas9 aspect of the CRISPR protocol, identifying a smaller version of the nuclease. The commonly used enzyme, sourced from Streptococcus pyogenes, was too big to fit into an adenovirus vector. But Cas9 from S. aureus is much shorter, the researchers found, and worked well delivered via viral vector to inhibit a cholesterol gene in mice. “What’s exciting about [saCas9] is it works really well in vivo,” Charlie Gersbach of Duke University, who was not involved in the study, told The Scientist in April.
Gerbach and colleagues made their own contributions to CRISPR that same month by hitching a histone acetyltransferase to Cas9 to create an epigenome editor. They had crippled the Cas9’s ability to chop DNA, using it instead as a homing device to get to the right spot in the genome and turn on a gene by acetylating the histone. Sigma Aldrich also developed a method to activate genes with CRISPR and made it available for customers in September, earning the firm a spot in The Scientist’s Top 10 Innovations of 2015.